Now, after finishing my master’s thesis, i would like to recap some experiences i made while working on it. Special emphasis will be put on the programming/data analysis aspect. The core of the thesis is a software called spectra mole, that combines Doppler spectra data from different active remote sensing instruments (Radar and Lidar) with the aim to observe vertical air motion inside clouds.
Language
This is kind of an endless discussion. Popular choices within the field, regarding interpreted/scripting/data analysis languages are: Matlab, IDL, R, Labview and Python
Endless arguments are held which one is superior to others, here are some reasons why I choose Python: it is open source (compared to Matlab, IDL and Labview) and a general purpose language (compared to R. It is not that much fun to do a plain Fourier transform with a statistics language). There are hundred thousands of tutorials available and nearly every question was already asked on stackoverflow.
Modularity and Unit Testing
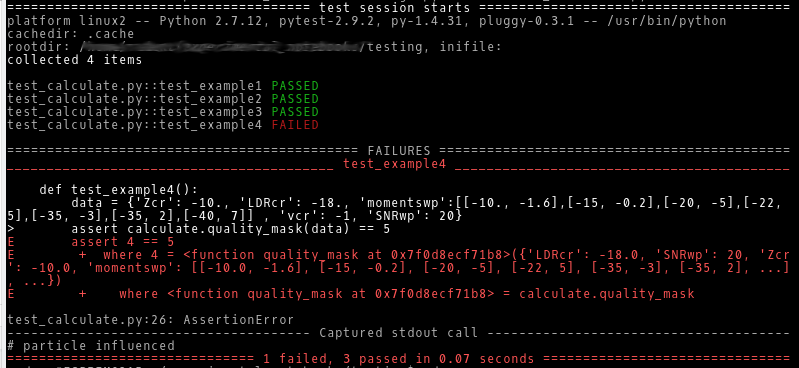
One major issue in writing scientific software is, that you have to assure that your calculations are correct. With increasing length and complexity of the algorithm, this becomes more and more difficult. The standard software development approach is to separate the program into small independent parts, that can be tested on their own.
The first version of my software was more or less divided into separate units, but without clear interfaces. So the units were quite entangled, which made testing nearly impossible. Refactoring was inevitable. Instead of generating one object and accessing the data at will, now a dictionary containing the data is passed from method to method (making this the “clear” interface). For each method a test case is implemented using pytest. This brought me a huge step forward in terms of reliability of the software.
Layers
Connected to the modularity topic is a layered design. This means that different levels of abstraction and processing steps are separated. For spectra mole this means, that data loading is separated from the processing and all auxiliary data is provided by separate modules. All post processing (mostly long term statistics) is done after a defined interface, which is in my case an output file.
Version Control/Source Code Management
Its a good idea to keep track of your source code history. The de-facto standard tool for that is git (besides maybe mercurial). One advantage is, that there are a bunch of tutorials and additional tools available. To name a few: github, bitbucket,GitKraken, etc.
Using version control, was a decision I made quite early, and it was a good one.
Visualization
Fast and easy plotting of the data can be quite helpful during development. The plotting routine is always a trade-off between flexibility, usability and quality (respectively beauty). The one tool for all purposes is hard to achieve.
Tracking
The general workflow is to write a chunk of analysis software, run it on some data and look at this data. If any quirks or points for improvement are found, they are implemented and the cycle starts again. Within this process it is necessary to track which version of the software was used on what data. This includes the settings used for each run. Such settings can be for example smoothing lengths, methods used, etc. Ideally they are saved in a machine readable text file using a markup language like json, toml or yaml.
For the software a source code management tool (see above) does this job quite well. But you should consider putting the text-files containing settings under version control, too. For the data itself i have found no solution, that works for me. Maybe saving the commit identifier of the used software version…
This leads to another point: the automation of the whole process.
Automation
Full tacking is possible, when the whole data modification/evaluation chain is automatized. This means, that all parameters and settings are stored in config files and all subroutines are called from one script. Even a simple shell script is enough. Of course this script and the config files should be version controlled.
When it becomes necessary, reprocessing can be done by going back to the old commit and running the script again.
Most of the progress in scientific programming is achieve in an iterative way, i.e. a small part of the code or a setting is changed and the data is reevaluated. Hence, it is in the responsibility of the programmer to “safe” his progress after each significant step.
Final thought
As most applied scientists, I received no extensive formal training on computer science and software development. It takes quite an effort to obtain these skills on your own and write software, that meets at least the most basic quality standards. One book that I can recommend in this context is “The Pragmatic Programmer: From Journeyman to Master” (ISBN: 978-0201616224).
Update 2019-12-14:
Googling the books title, you will likely find a downloadable version of the book.